Molecular analysis via PCR & gel electrophoresis protocols now complement phenotypic observations with Fast Plants
New classroom PCR and gel electrophoresis protocols were introduced for use with Fast Plants this 2018/19 school year. With these new biotechnology options, learners can see firsthand the connection between an easily observed phenotypic trait and its underlying genotype. Whether you choose MiniPCR, Carolina Biological or any of the other biotech suppliers, the molecular door is now open for using readily-available Fast Plants stocks to investigate the genetics that underpin observed Mendelian inheritance patterns.
As an introduction to this exciting addition to the Fast Plants repertoire, the MiniPCR team wrote the following guest post about their freshly-released protocol and classroom kit.
Wisconsin Fast Plants® have been a staple of Biology classrooms for decades. With short generation times, high productivity, and many distinct visual phenotypes, generations of students have used Fast Plants® to study a variety of topics ranging from energy flow through ecosystems to Mendelian genetics. But as molecular techniques such as PCR and gel electrophoresis have entered high school classrooms, there has been a gap for teachers who would like to link classical genetics breeding with more modern approaches. The miniPCR™ Plant Genetics Lab looks to bridge that gap. Students can now add molecular approaches to their Wisconsin Fast Plant investigations, linking a clearly observable phenotype to its underlying genetic cause.
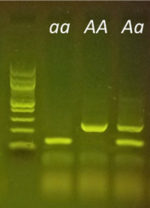
 Purple versus non-purple stem color is a commonly used trait in classrooms, especially when investigating Mendelian genetics. Based on recently published research[1], we now know that Anthocyaninless, the gene responsible for the two phenotypes, is a gene that codes for the production of the enzyme Dihydroflavonol 4-reductase (DFR). DFR is an integral enzyme in the production of the purple pigment, anthocyanin. The recessive green allele (anl, or a) contains a 354 base pair insertion mutation. Those extra 354 base pairs knock out the function of the resulting protein in the “a” allele, while the purple allele (ANL, or A) allele produces a normal, functional DFR protein. This difference in the genetic sequence is easily detectable using PCR and gel electrophoresis. Students can now predict a plant’s genotype based on its phenotype and then test their predictions right in the classroom using molecular techniques.
Purple versus non-purple stem color is a commonly used trait in classrooms, especially when investigating Mendelian genetics. Based on recently published research[1], we now know that Anthocyaninless, the gene responsible for the two phenotypes, is a gene that codes for the production of the enzyme Dihydroflavonol 4-reductase (DFR). DFR is an integral enzyme in the production of the purple pigment, anthocyanin. The recessive green allele (anl, or a) contains a 354 base pair insertion mutation. Those extra 354 base pairs knock out the function of the resulting protein in the “a” allele, while the purple allele (ANL, or A) allele produces a normal, functional DFR protein. This difference in the genetic sequence is easily detectable using PCR and gel electrophoresis. Students can now predict a plant’s genotype based on its phenotype and then test their predictions right in the classroom using molecular techniques.
A quick and classic approach to investigate the Anthocyaninless allele is to germinate F2 seeds derived from an original AA x aa parental cross. Students can then see if seeds match the expected 3:1 phenotypic ratio according to Mendelian predictions and test their data using the chi-squared statistic. With the miniPCR Plant Genetics Lab, students can take the approach one step further. Students can now test whether plants also adhere to the 1:2:1 genotypic ratio as predicted by Mendel, all with a simple DNA extraction, PCR, and gel electrophoresis protocol that can be easily completed in two class periods.
An exciting aspect of this lab, however, is that the it can be employed using several different approaches. Wisconsin Fast Plants® have been bred to have short generation times with prolific seed production, so multiple generations can be grown by students in a class over a period of several weeks. This allows teachers to take a more inquiry, student-centered approach to learning genetics. Have students grow the parental strains, perform the appropriate crosses and create their own F1 and F2 plants. Students can then predict both the genotypic and phenotypic ratios of each generation. By testing the genotypes of parental, F1, and F2 plants, students can actually visualize the inheritance of alleles over time.
 Or give students seeds of unknown lineage and challenge them to determine the genotype through breeding. By observing the phenotypes of the offspring, students predict the genotypes of the parents. With the miniPCR Plant Genetics Lab, students can save samples from the parental plants and confirm if their analysis of the offspring was correct – effectively testing their predictions and seeing real and authentic applications of biotechnology in the classroom.
Or give students seeds of unknown lineage and challenge them to determine the genotype through breeding. By observing the phenotypes of the offspring, students predict the genotypes of the parents. With the miniPCR Plant Genetics Lab, students can save samples from the parental plants and confirm if their analysis of the offspring was correct – effectively testing their predictions and seeing real and authentic applications of biotechnology in the classroom.
The miniPCR machine was invented as an affordable biotechnology tool that could put real science in the hands of students, increasing both access and engagement. We are excited to introduce this new lab, helping to further extend the many ways Wisconsin Fast Plants can be used in the classroom. Being able to show that molecular techniques are not a separate endeavor, but an extension to the science that students are already doing is an invaluable way of connecting advanced techniques to basic biology. Join the DNA revolution!
[1] Wendell, D.L. Vaziri, A. Shergill, G. (2016) The Gene Encoding Dihydroflavonol 4-Reductase Is a Candidate for the anthocyaninless Locus of Rapid Cycling Brassica rapa (Fast Plants Type) PLoS One, 11(8)
Links to PCR & gel electrophoresis kits, equipment, and Fast Plants seeds:
MiniPCR kit: http://tinyurl.com/y2886o2v
MiniPCR equipment: https://www.minipcr.com/products/
Carolina Biological Product Page for F2 Non-Purple stem seeds
Additional References
Teaching Human Genetics with Mustard: Rapid Cycling Brassica rapa (Fast Plants Type) as a Model for Human Genetics in the Classroom Laboratory by Douglas L. Wendell* and Dawn Pickard; Paul Williams, Monitoring Editor: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1885900/